Automated SARS-CoV-2 Nanopore Sequencing Analysis for Pandemic-Scale Diagnostics
Published: 2025-07-02
The COVID-19 pandemic exposed key bottlenecks in the ability of health systems to rapidly process and interpret pathogen genomic data. Although command-line bioinformatics pipelines enabled automation and high-throughput SARS-CoV-2 genome analysis, their reliance on programming expertise limited their adoption in many diagnostic and clinical labs. As sequencing data became central to understanding viral evolution, and guiding public health interventions, there emerged a critical need for tools that were powerful, yet accessible, to users without coding backgrounds.
Oxford Nanopore Technology (ONT) has gained recognition for its capacity to deliver long-read sequencing data cost-effectively, and with short turnaround times. This makes it suitable for real-time outbreak tracking. However, many ONT data analysis pipelines (e.g. Artic, NanoCoV19, Viralrecon) were exclusively operated using command-line environments. Bridging this usability gap, graphical user interface (GUI) software, such as Geneious Prime, emerged as a promising platform to empower lab personnel with limited bioinformatics training to efficiently perform complex sequencing analyses.
The work by Cumlin et al. (2024) aimed to develop automated, plugin-based workflows that would allow users to perform SARS-CoV-2 ONT sequencing analysis, including consensus generation and phylogenetic assignment, without requiring programming skills. The workflows are designed to be modular, cross-platform, and adaptable for future pathogen outbreaks.
Two validated workflows were developed as part of this work:
- SARS-CoV-2_Assembly_Basic: Performs trimming, alignment, consensus sequence generation, and annotation of ONT data.
- SARS-CoV-2_Assembly_WrapperPlugins: Enhances the SARS-CoV-2_Assembly_Basic workflow by integrating Pangolin and Nextclade as plugins for real-time lineage assignment.
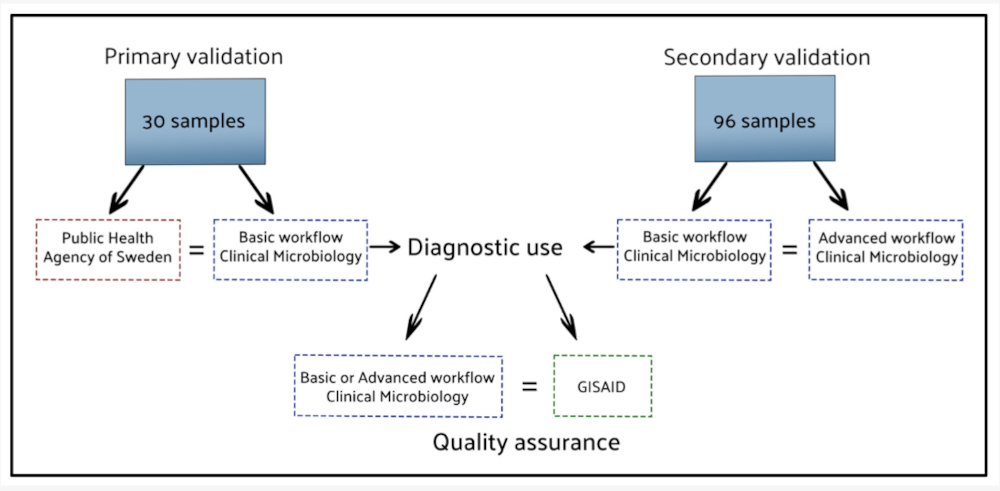
The SARS-CoV-2_Assembly_Basic workflow was previously validated against the method used by the Public Health Agency of Sweden by analysing 30 sequences in parallel. Validation of the SARS-CoV-2_Assembly_WrapperPlugins workflow was performed using 96 clinical samples, and the results were benchmarked against results from the web interfaces of Pangolin and Nextclade. This benchmarking showed that, when the Pangolin and Nextclade are intergrated as part of the SARS-CoV-2_Assembly_Basic workflow, the results were both accurate and fully concordant with their respective web interfaces. The automated workflows reduced hands-on time, and simplified analysis across Linux, macOS, and Windows operating systems. Visual inspection features within Geneious Prime enabled mutation tracking and quality control, with real-world use in Sweden’s national surveillance programme.
The development and deployment of GUI-based, plugin-supported workflows for SARS-CoV-2 sequencing data addresses a pressing gap in pandemic preparedness infrastructure. Their adoption within Uppsala University Hospital, and integration into Sweden’s national surveillance program (see e.g. Martinell et al. (2022)), illustrates their utility at scale. Beyond SARS-CoV-2, the modular design allows adaptation to other pathogens by altering primers and reference genomes, positioning this toolset as a flexible resource for future outbreaks, mutation surveillance, and antimicrobial resistance tracking. These workflows exemplify how democratised data analysis, via intuitive platforms, can advance the goals of data platforms and pathogen portals; enhancing global collaboration, reducing diagnostic inequity, and enabling timely public health action.
Data
- Workflows available on GitHub.
- Sequence test data for workflow validation available at NCBI (Accession number PRJNA1048178).
- Videos describing how to set up and run the workflows are available on the research group’s YouTube channel.
Informed consent was obtained from all subjects involved in the study. All raw data processing was conducted in a clinical setting for method development. This study used only existing sequence data, with all consensus files having been made publicly available beforehand.
Article
DOI: 10.3390/ijms25126645
Cumlin, T., Karlsson, I., Haars, J., Rosengren, M., Lennerstrand, J., Pimushyna, M., Feuk, L., Ladenvall, C., & Kaden, R. (2024). From SARS-CoV-2 to Global Preparedness: A Graphical Interface for Standardised High-Throughput Bioinformatics Analysis in Pandemic Scenarios and Surveillance of Drug Resistance. International Journal of Molecular Sciences, 25(12), 6645.
Funding
This research was funded by: (1) Wallenberg foundation (KAW 2020.0241) ‘High throughput SARS-CoV-2 variants surveillance’ (Lars Feuk, Rene Kaden) and (2) Gullstrand foundation (ALF-938076): ‘Development, evaluation and implementation of molecular epidemiological source tracing methods and variant surveillance’ (Rene Kaden).