New data highlight on GUI-based workflows for SARS-CoV-2 Nanopore sequencing analysis
Published: 2025-07-02
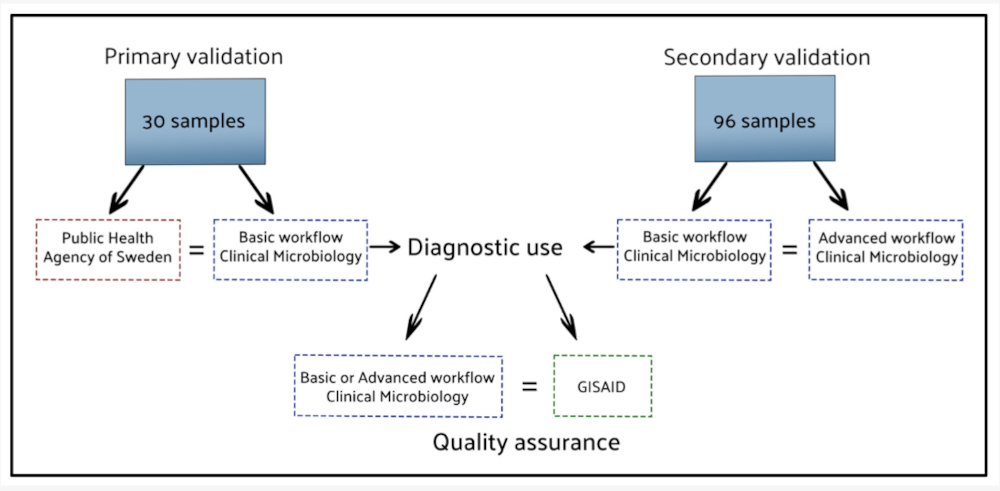
Our data highlights showcase research that shares data and/or other research outputs in as FAIR and Open a manner as possible. The latest data highlight, entitled ‘Automated GUI-based workflows improve accessibility of SARS-CoV-2 Nanopore data analysis’, shows examples of tools that can be repurposed for multiple pathogens, thus contributing to future pandemic preparedness. Here, researchers at Uppsala University Hospital developed and validated two automated workflows within Geneious Prime, a graphical software platform, to streamline SARS-CoV-2 Oxford Nanopore sequencing analysis.
In particular, Cumlin et al., 2024, provides a practical solution for labs with limited expertise in coding. The workflows perform trimming, alignment, consensus sequence generation, and (in the advanced version) phylogenetic lineage assignment, using Pangolin and Nextclade as integrated plugins.
These user-friendly workflows are already deployed in Sweden’s national SARS-CoV-2 surveillance program (see e.g. Martinell et al. (2022)), and have been validated across Linux, macOS, and Windows operating systems. Importantly, they have reduced analysis time while preserving diagnostic accuracy.
Validation data and tools are openly available via GitHub, and the Sequence Read Archive (accession number: PRJNA1048178), ensuring others can adopt or adapt the workflows for different pathogens and outbreak scenarios.
To find out more about the study, please read the data highlight.
If you are interested in having your work in the areas of pandemic preparedness and/or infectious disease promoted on the portal (e.g. with a data highlight like this one), please contact us.