New data highlight on Host range expansion in Giardia duodenalis linked to Muller's Ratchet
Published: 2026-01-29
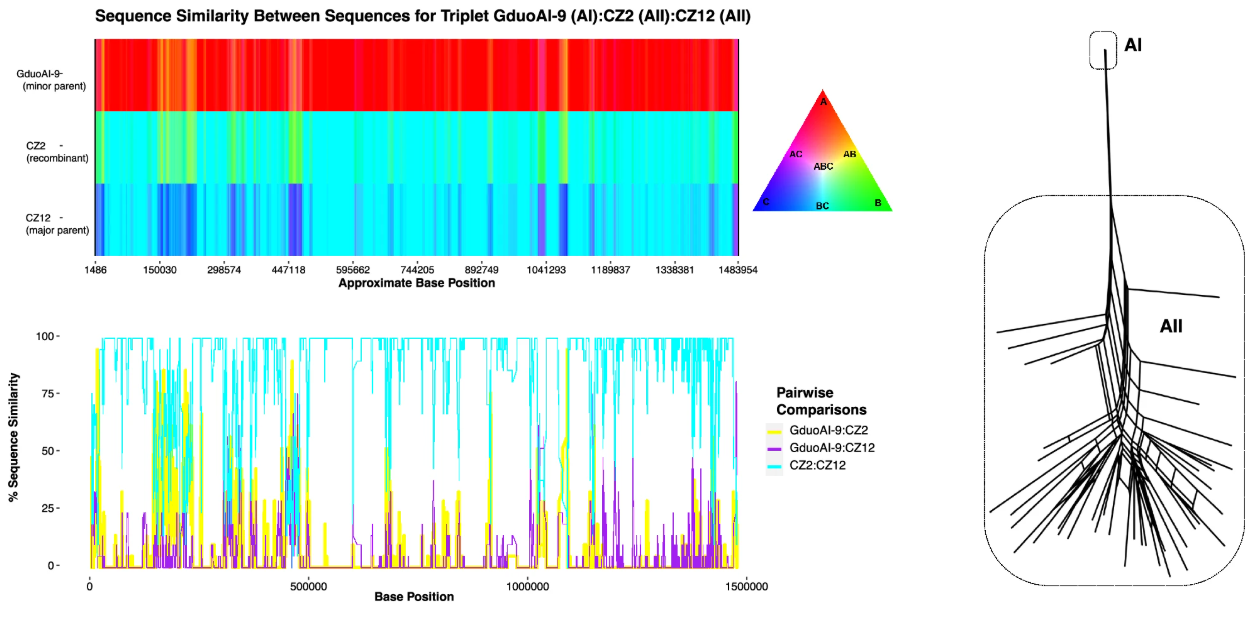
Our latest data highlight ‘Host range expansion of asexual parasite can be explained by loss of adaptions in Muller’s Ratchet’ is focused on the work of Tichkule et al.(2025), which explores the evolutionary genomics of Giardia duodenalis. Tichkule et al. (2025) provides a mechanistic explanation for how an asexual lineage of Giardia broadens its host range, despite the expected constraints of long-term asexuality. Tichkule et al. (2025) have openly shared sequencing data in the NCBI Sequence Read Archive (SRA) under the BioProject PRJNA1110996.
The study combines comparative population genomics and evolutionary modeling to distinguish sexual (AII) and asexual (AI) lineages. Using whole-genome SNP profiling of 101 isolates across diverse hosts excluding the 6 mixed infections, Tichkule et al. (2025) revealed that AI lacks evidence of recombination, accumulates deleterious mutations, and exhibits disrupted host-specific adaptations. In contrast, the sexual AII lineage shows recombination signals and more constrained host specificity. Analyses of linkage disequilibrium, mutation accumulation, and phylogenomic clustering support the conclusion that Muller’s Ratchet drives genome evolution in the AI lineage, paradoxically enabling temporary host-range expansion.
This research offers a new perspective for parasite biology and evolutionary genomics, demonstrating how the accumulation of mutations in asexual lineages can reshape adaptations and allow transient expansion into new hosts. Researchers studying host-pathogen evolution, population genetics of asexual organisms, or mutational processes in non-recombining genomes will find this resource particularly valuable.
To find out more about the study, please read the data highlight.
If you are interested in having your work in the areas of pandemic preparedness and/or infectious disease promoted on the portal, please contact the portal team.